16 May 2023
We are happy to announce that MDAnalysis is hosting two GSoC students this year - @xhgchen and @marinegor. MDAnalysis has been accepted as its own organization with GSoC for the fourth year running, and we are grateful to Google for granting us the opportunity to get started on two very exciting GSoC projects!
Xu Hong Chen: Add calculations for self-diffusivity and viscosity

Transport properties are values defining mass, momentum, heat, or charge transfer and are vital to biomolecular research and chemical engineering. MDAnalysis currently provides little support for these calculations and adding them to MDAnalysis would greatly benefit researchers and engineers. Xu Hong will implement a class to calculate self-diffusivity via the Green-Kubo method and utility functions to compute shear viscosity via the Einstein and Green-Kubo methods. He will test and revise his algorithms rigorously to ensure they are accurate and efficient. To maximize user accessibility, Xu Hong will write clear and understandable documentation linked to the best practices in the literature. Xu Hong’s project page can be found here.
Xu Hong is an incoming computer science student at the University of British Columbia and a biochemistry graduate from the University of Alberta. He has 2 years of experience in molecular dynamics from his research on the molecular mechanism of Hsp90 in the Spyracopoulos Lab. He is interested in scientific software, high-performance computing, computational research, and open-source software. To rest and relax, Xu Hong enjoys playing piano and listening to Chopin, Schubert, and Bach.
Xu Hong is on GitHub as @xhgchen and on LinkedIn as Xu Hong Chen. He will be sharing his experience on his blog.
Egor Marin: Implementation of parallel analysis in MDAnalysis

Even though MDAnalysis (since 2.0) allows for serialization of almost all fundamental components, it still lacks a seamless way to run analysis of trajectories in a parallel fashion. For his GSoC project, Egor will be implementing a parallel-ready backend for MDAnalysis, hoping that it will increase the speed and accessibility of large-scale molecular dynamics analysis.
Egor will hopefully graduate this year fom the PhD program of the University of Groningen (Netherlands). His main expertise lies in structural biology, as can be seen by his work during his bachelor’s and master’s. Apart from that, he does a lot of computer administration work, and enjoys occasional bouldering and snowboarding.
You can find Egor on GitHub and Twitter. You can follow Egor’s GSoC work throughout the summer on his blog.
— @hmacdope @orionarcher @yuxuanzhuang @RMeli @orbeckst @richardjgowers @ianmkenney @IAlibay @jennaswa (mentors and org admins)
20 Apr 2023
We’re happy to announce that the SolvationAnalysis Python package is now published in the Journal of Open Source Software (JOSS):
Orion Archer Cohen, Hugo Macdermott-Opeskin, Lauren Lee, Tingzheng Hou, Kara D. Fong, Ryan Kingsbury, Jingyang Wang, and Kristin A. Persson, (2023). SolvationAnalysis: A Python toolkit for understanding liquid solvation structure in classical molecular dynamics simulations. Journal of Open Source Software, 8(84), 5183, https://doi.org/10.21105/joss.05183
Originally developed by @orionarcher as a GSoC 2021 project, SolvationAnalysis builds on MDAnalysis and pandas to make analyzing solvation structure much, much easier. With a few lines of code, you can extract key solvation properties from any molecular dynamics simulation. With a few more, you can visualize solvation trends within and between different solutions. Further, SolvationAnalysis is designed with extensibility in mind, exposing a core representation of solvation that users can use for brand new analyses. To get started with using SolvationAnalysis in your workflow, check out the documentation and tutorials.

24 Mar 2023

MDAnalysis is applying for Google Season of Docs (GSoD) 2023. In this program, Google is sponsoring technical writers to work with open source projects in the area of documentation. Below is our project proposal we’ve submitted for this year’s program.
Update, Cleanup, and Simplification of Documentation and Learning Resources
About your organization
MDAnalysis (current version 2.4.2, first released in January 2008) is an object-oriented Python library for temporal and structural analysis of molecular dynamics (MD) simulation data. MD simulations of biological molecules are an important tool for elucidating the relationship between molecular structure and physiological function. With thousands of users world-wide, MDAnalysis is one of the most popular packages for analyzing computer simulations of many-body systems at the molecular scale, spanning use cases from interactions of drugs with proteins to novel materials. As MDAnalysis can read and write simulation data in 30 coordinate file formats, it enables users to write portable code that is usable in virtually all biomolecular simulation communities. MDAnalysis forms the foundation of many other packages and is currently used by more than 20 data visualization, analysis, and molecular modeling tools. All MDAnalysis code and teaching materials are available under open source licenses, and the library itself is published under the GNU General Public License, version 2 or any later versions (GPLv2+).
About your project
Your project’s problem
MDAnalysis has an ever-expanding international community of users and developers. Its success can be attributed not only to its high-quality codebase, but also to its extensive learning resources and thorough documentation. MDAnalysis’s participation in Google Season of Docs (GSoD) 2019-2020 was a quantum leap for our project, as a technical writer created the User Guide with the Quick Start Guide, which have become the primary entry points for new users. The GSoD 2019-2020 work also catalyzed development of tutorial and online workshop materials. Although these materials are publicly available under open source licenses and are valuable resources for users, there is currently a considerable amount of overlap between MDAnalysis’s existing learning resources (main website, user guide, docs, GitHub wiki), making it difficult for self-learners to find the information they need (Figure 1). Specifically, MDAnalysis resources (1) are duplicated across multiple hubs of information, (2) are organized such that new users stumble across developer-focused content early on, and (3) contain outdated tutorials and examples corresponding to older versions of the library.
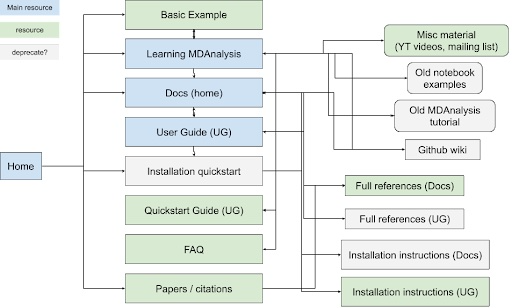 |
|
Figure 1: Map of current MDAnalysis learning resources and how they link to each other. Blue boxes indicate main hubs of material that also link to other resources, while green boxes correspond to simple resources. Resources inside gray boxes could be deprecated and their contents moved to other sites, if they are not already duplicated elsewhere. |
For example, installation instructions can be accessed from the main website, user guide, docs, and GitHub wiki. As this is the first documentation new MDAnalysis users are likely referencing, it can cause confusion and overwhelm newcomers when they are directed to multiple places through a series of hyperlinks. Similarly, our application programming interface (API) guide is accessible from the same location as the user guide, potentially directing new users to resources intended for developers. Through this project, we intend to more clearly delineate between API documentation and the user guide to improve junior developers’ experiences with navigating our documentation. Furthermore, we expect that this will make it easier for mentees under programs such as Google Summer of Code (GSoC) or Outreachy to transition towards becoming MDAnalysis developers.
We also expect that improving user experience with MDAnalysis learning resources will lower the barriers to adoption of MDAnalysis compared to other similar packages (e.g., GROMACS or MDTraj). Funded by a 2-year Essential Open Source Software for Science (EOSS) round 5 grant awarded to MDAnalysis by the Chan Zuckerberg Initiative (CZI), we will offer three online teaching workshops per year. Having a more succinct user guide, easy-to-find tutorials, and updated examples will enhance the user experience for new MDAnalysis users.
Our core developers currently spend a considerable amount of time monitoring questions asked on our user mailing list. Many of these inquiries can be resolved by directing users to existing content on the main website, in the user guide, or in the docs. The reorganization of resources proposed in this project would thus provide users with a faster and more efficient way to access relevant information without relying on core developers to respond to the mailing list.
Your project’s scope
Through this project, we propose a cleanup of the main website and additional MDAnalysis learning resources to guide users through a streamlined workflow (Figure 2). The main body of each site (main website, user guide, docs) would guide the user through the user process by limiting available choices and allowing quick jumping between the main hubs of information. The GitHub wiki would be devoid of user content and would instead become a developer-focused resource.
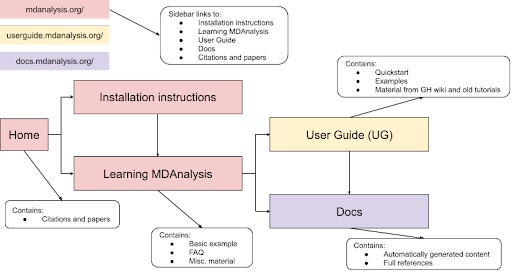 |
|
Figure 2: Proposed flow of MDAnalysis learning resources to ease user navigation. |
In particular, the MDAnalysis project will:
- Remove duplication between the main website, user guide, docs, and the GitHub Wiki by defining the role of (and which content should be included in) each resource
- Identify and update any outdated material according to recent code releases
- Integrate existing workshop materials and tutorials into the user guide
- Move non-automatically generated content from docs into the user guide
- Differentiate between API guide and user guide
- Merge installation instructions across all resources
- Identify and remove old examples, outdated tutorials, and deadlinks
- Transition the GitHub Wiki into a developer-focused resource to prevent overwhelming MDAnalysis users with unnecessary details while guiding advanced users towards becoming developers (through bridges in the user guide)
- Address some issues compiled on the user guide GitHub repository
Work that is out-of-scope for this project:
- This project will not generate new learning resources; rather, it will reorganize existing content to enable users to more easily locate and access resources.
- This project will not make any changes to the MDAnalysis mailing list, and will not use the mailing list content to compile users’ frequently asked questions (FAQ) into an updated FAQ page.
We have identified a strong technical writing candidate, Dr. Patricio Barletta (@pgbarletta), for this project, and we estimate that this work will take 7.5 months to complete. Dr. Barletta is currently a junior developer contracted to improve the organization and content specifically of MDAnalysis’s workshop teaching materials, sponsored by NumFOCUS through a Small Development Grant (150 hours, $3,600).
The project is set to affect multiple repositories and involves general restructuring of many resources, thus we anticipate a high level of involvement from the core developer team. Four MDAnalysis core developers - Dr. Micaela Matta (@micaela-matta), Dr. Irfan Alibay (@IAlibay), Prof. Oliver Beckstein (@orbeckst), and Lily Wang (@lilyminium) - will be responsible for reviewing pull requests, assisting with site search and indexing tasks, mentoring the technical writer, and providing other necessary support to the GSoD program. The MDAnalysis program, community, and outreach manager, Dr. Jenna Swarthout Goddard (@jennaswa), will serve as the organization administrator for the GSoD program.
Measuring your project’s success
We will measure the success of our project according to the following metrics:
-
Reduced number of docs-related issues on GitHub repository. We currently have 33 open docs-related issues on the MDAnalysis GitHub repository and another 51 issues on the user guide repository. In addition to specifically addressing these issues as an objective of this project, we anticipate the update, cleanup, and reorganization of our docs will close at least 7% of these outstanding issues (e.g., those related to duplication or outdated examples).
-
Increased number of visits to MDAnalysis learning resources. We regularly (and publicly) track web analytics for our main website using GoatCounter (currently >50,000 unique visits per month). We will thus track whether the number of visits to the website, as well as to specific documentation (e.g., user guide, tutorials, etc.), increase once our improved documentation is published. We aim to increase visits to current tutorials and learning resources by 30%.
-
Increased number of MDAnalysis installations and citations. A primary objective of this project is to enhance the user experience for newcomers to MDAnalysis and ultimately encourage continued growth in our user community. New releases are downloaded more than 27,000 times per month (according to condastats and PyPI Stats over the last 12 months) and the academic papers describing MDAnalysis are cited over 2,500 times (Source: Google Scholar). A major milestone for this project will therefore be an increase in the number of new MDAnalysis installations, measured by >27,000 downloads per release. As a measure of sustained use of MDAnalysis, potentially indicating ease of use compared to similar packages in the academic community, we will monitor whether there is also an increase in the rate of MDAnalysis citations per year (Figure 3).
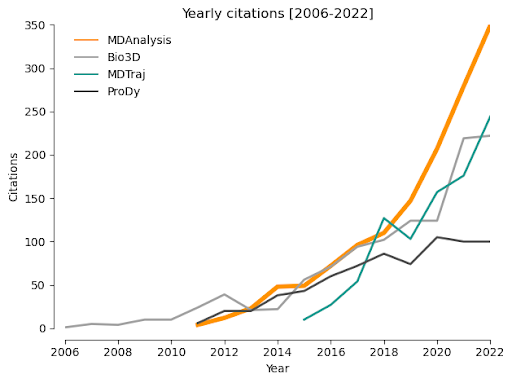 |
|
Figure 3: Yearly citations for MDAnalysis compared to other similar packages between 2006 and 2022 (Source: Scopus). |
-
Increased number of contributions to MDAnalysis. We anticipate that clear separation of information regarding contributing in the user guide and the API guide, as well as general improvements to the overall documentation, will ease the transition for users to developers. Concurrent with our participation in GSoD 2019-2020, we observed a more than 2.5-fold increase in pull requests - and specifically in new contributor pull requests - between 2019 and 2020. We thus expect our number of repository forks, commits, and pull requests to rise by at least 20%.
Finally, MDAnalysis is participating as an early adopter of an initiative of a NumFOCUS and Bitergia partnership to establish and monitor community metrics in line with the Community Health Analytics in Open Source Software (CHAOSS) project. Dashboard setup and identification of relevant metrics are currently under development. Incorporating quantitative metrics specifically measuring community health (e.g., what contributions are made and by who), diversity, equity, and inclusion (DEI), and community growth will be especially important in evaluating the impacts of improving our documentation.
Timeline
The project itself will take approximately 7.5 months to complete. Once the tech writer is hired, we will spend a month on tech writer orientation and identification of outdated material, then move onto the integration and updating of existing materials, and spend the last month addressing user guide issues in our GitHub repository.
| Dates |
Action Items |
| April |
Orientation and identification of outdated material. |
| May |
Integrate existing documentation into user guide and remove duplication between main website, user guide, and docs. |
| June |
Create a separate API guide, distinct from the user guide. |
| July |
Merge installation instructions from main website, user guide, docs, and GitHub wiki. Remove old tutorials, old examples, and deadlinks. |
| August |
Remove user content from GitHub wiki, incorporate this content into the user guide, and shift the wiki towards developer-focused content. |
| September |
Start addressing issues from the GitHub user guide repository. |
| October |
Continue addressing issues from the GitHub user guide repository. |
| November |
Project completion. |
Project budget
| Budget Item |
Amount |
Running Total |
Notes/Justification |
| Technical writer time |
$7,000 |
$7,000 |
200 hours (6.25 hours/week for 32 weeks) at a rate of $35/hour |
| Mentor stipends |
$2,000 |
$9,000 |
4 volunteer stipends at $500 each |
| Graphic designer |
$1,000 |
$10,000 |
Development of illustrations for inclusion in updated user guide to accompany tutorials and illustrate new features (i.e., MDAKits) |
| Swag |
$200 |
$10,200 |
T-shirts, stickers, etc. |
| TOTAL |
|
$10,200 |
|
Previous experience with technical writers or documentation
MDAnalysis previously worked with a technical writer through the GSoD 2019-2020 program. The technical writer, @lilyminium (MDAnalysis core developer since 2020), not only updated the user guide, but also improved overall document appearance, fixed inconsistencies between materials, contributed code to the MDAnalysis library, and supported users. As @lilyminium was herself an MDAnalysis user, she was incredibly successful at independently generating new content and interacting with users to identify ongoing needs in the existing documentation. However, as the work we have proposed for GSoD 2023 involves restructuring existing material as opposed to generating new content, we would like this year’s candidate to approach the project from the perspective of a relatively new MDAnalysis user. Our GSoD 2023 candidate, @pgbarletta, has been working with the project since September 2022 on a NumFOCUS Small Development Grant aimed at improving and restructuring tutorial materials for future MDAnalysis workshops. As a newcomer to the project, he was quickly able to identify gaps and redundancies in the user guide and overall online documentation. His proposed improvements and modifications form the core of this GSoD proposal.
Previous participation in Google Season of Docs, Google Summer of Code or others
In addition to participation in GSoD 2019-2020 as described above, MDAnalysis has mentored 13 students through GSoC since 2016 (1-3 students annually) and 1 student through Outreachy 2022. Together, these initiatives have provided mentorship and support to early-stage open source developers, as well as encouraged progress on important issues and attracted long-term contributors. Several past GSoD, GSoC, and Outreachy participants are now core developers or ongoing contributors to the MDAnalysis library. In fact, 10 participants mentored through GSoD, GSoC, and Outreachy remain involved with the MDAnalysis project, with 4 of them serving as core developers and an additional 2 volunteering as mentors in this year’s GSoC program. Furthermore, MDAnalysis was awarded two CZI EOSS (rounds 4 and 5) grants. CZI EOSS4 has resulted in the establishment of the MDAKits, an ecosystem of downstream packages contributed by users of the main library. CZI EOSS5 enabled MDAnalysis to hire a dedicated project manager (Dr. Swarthout Goddard) to enhance the scope of MDAnalysis’s outreach, mentoring, and dissemination efforts. Dr. Swarthout Goddard will be providing administrative support throughout GSoD.




