11 Feb 2025

As we enter the new year, we at MDAnalysis are still reflecting on and grateful for the work of our contributors in 2024. Namely, we would like to recognize the completion of Luna Morrow’s (@lunamorrow) Google Summer of Code (GSoC) project with MDAnalysis, Extend MDAnalysis Interoperability with OpenBabel — read more about this incredibly useful enhancement of MDAnalysis’s interoperability with other packages in the field in Luna’s blog post.
We have enjoyed watching Luna navigate the full cycle of planning, designing, implementing, testing, and documenting a software solution and look forward to seeing what she accomplishes next!
We also thank Google for supporting this project and continuing to support MDAnalysis as a GSoC organization since 2016.
Looking ahead to GSoC 2025
The MDAnalysis community remains enthusiastic about continuing to participate in the GSoC program. Given that there is an application process for organizations — such as MDAnalysis — to participate, there is no guarantee that MDAnalysis will be selected by Google as a participating GSoC organization. Once we know more, we will share updates on our #gsoc Discord (join using this invite link) and #GSoC Discussions GitHub Discussions channels.
In the meantime, we welcome you to start learning MDAnalysis and familiarizing yourself with the project through our introductory videos. To dive into the code base, check out the MDAnalysis User Guide; we suggest starting by installing the MDAnalysis package and working through the Quick Start Guide. Once you are a bit familiar with the MDAnalysis package, you can look at the User Guide sections explaining how to contribute.
Please note that in addition to code contributions, we highly value participating in the MDAnalysis community in other ways, including submitting general feedback and issues via the GitHub issue tracker and/or engaging in discussions on the MDAnalysis Discord server (join using this invite link) and GitHub Discussions forum.
— @cbouy @hmacdope @IAlibay @jennaswa @richardjgowers @xhgchen @yuxuanzhuang (GSoC 2024 mentors and org admins)
09 Feb 2025
I can’t believe I am at the end of GSoC! This past 6 months has absolutely flown by, but I have learnt so much. I am really grateful for the amazing support I have had along the way from my mentors Hugo ( @hmacdope ), Cédric ( @cbouy ) and Xu ( @xhgchen ). My body of work can be located on the MDAnalysis GitHub at mda-openbabel-converter.
Why a converter
Direct interoperability between molecular dynamics software is critical for enabling collaboration, data transfer and straightforward use by scientists. OpenBabel is a popular toolbox for chemical molecular modelling research as it enables searching, conversions, analysis and data storage. The ability to interconvert chemical file formats with OpenBabel, in particular, opens up the ability to utilise and work with other packages, as OpenBabel enables input and writing of over 100 chemical data file formats. Therefore, enabling an MDAnalysis Universe to be inter-converted with an OpenBabel OBMol would greatly increase the data formats available to MDAnalysis. Furthermore, it would encourage greater adoption of MDAnalysis as an “all-in-one” package for molecular dynamics analysis.
From OpenBabel to MDAnalysis
The first aim of my GSoC project was to convert OpenBabel OBMols to MDAnalysis Universes. This required the extraction of both molecule and atom data, alongside their 3D positions. I hit many roadblocks in this section, with the two most difficult being the installation of OpenBabel 3.1.1.1 Python bindings and understanding some of the more complex OpenBabel API. Around half of the coding period was spent writing the two classes for converting the atom attributes and positions respectively. Time was also committed to developing tests for both classes.
First, I created the OpenBabelParser Class (converts the atom attributes) and some basic unit tests for it in #12. Once this PR was opened, it revealed that the CI was not correctly setup. With additional help from Irfan ( @IAlibay ), this was corrected in #13. Next, I made the OpenBabelReader Class (converts the atom positions) and a whole suite of tests for it in #16, and then applied some of these ‘extended’ test conditions to the OpenBabelParser in #18. During this time some issues were found with compatibility between, and accessing of, attributes from OpenBabel. I opened an issue on the OpenBabel GitHub to obtain assistance (see here) and also made a issue to flag an attribute that was proving difficult but will need to be supported later in #17.
Documentation
Once the OpenBabelParser and OpenBabelReader were developed, I setup ReadTheDocs #19 and wrote documentation for these two classes #20. The documentation is readily available on ReadTheDocs here. The documentation is functional but currently quite sparse, I have created an issue to improve the landing page and getting started page, once functionality is complete, at #21.
From MDAnalysis to OpenBabel
The next stage of this this project was to implement the OpenBabelConverter Class to convert an MDAnalysis Universe to OpenBabel OBMol. Unfortunately, I have run out of time to implement this class during the allotted time for GSoC. There is an open issue detailing this class at #22, which I will be expanding on shortly. While my time with GSoC has ended, I am keen to complete this project in my own time and continue on with MDAnalysis as a developer.
Next Steps
I will be finishing the OpenBabelConverter, including tests and documentation. The next steps will then be to add attributes that were left for later, further development of the documentation with worked examples and deploying a release of this package. I would also like to integrate this converter into the MDAnalysis package so that it can be used alongside the other converters, as this would increase its visibility and usage.
What can we do now?
With what I currently implemented at the end of the GSoC coding period, users can very easily convert an OBMol to an MDAnalysis universe.
An example of how to use the converter to convert an OBMol to an MDAnalysis universe is shown below:
from openbabel import openbabel as ob
import MDAnalysis as mda
obconversion = ob.OBConversion()
obconversion.SetInFormat("pdb")
mol = ob.OBMol()
obconversion.ReadFile(mol, "1crn.pdb")
u = mda.Universe(mol)
What I’ve Learned
Things I have learned during GSoC include:
- the importance of good documentation
- how to develop the ‘backbone’ of a python package so it can be installed, tested and used
- developing tests with pytest
- managing myself and being able to make decisions about when something is out of scope or unviable to implement
- how to use git and GitHub
- developing classes that inherit from classes that were not developed by me
My experience
I have had an amazing time during GSoC. I had a lot of support and felt very welcomed and encouraged. I can’t wait for this converter to be up and running, so that the community can benefit from it. I am also really grateful for the experience and technical growth GSoC granted me, as I know it will be incredibly beneficial for my future in this field.
12 Dec 2024

We are thrilled to announce that MDAnalysis has been awarded a Small Development Grant by NumFocus to enhance scientific molecular rendering with MolecularNodes in 2025. This initiative is a collaborative effort between Yuxuan Zhuang and Brady Johnston.
MolecularNodes facilitates the seamless import and visualization of structural biology data within Blender, leveraging Blender’s industry-leading visualization and animation tools. Molecular Nodes has garnered widespread excitement among scientists for its ability to create stunning and informative molecular visualizations. The current version of Molecular Nodes has an under-developed scripting interface, inhibiting the potential for automated molecular rendering. Our project aims to address these limtiations by developing a robust API, enabling users to render molecular structures with straightforward and customizable code.
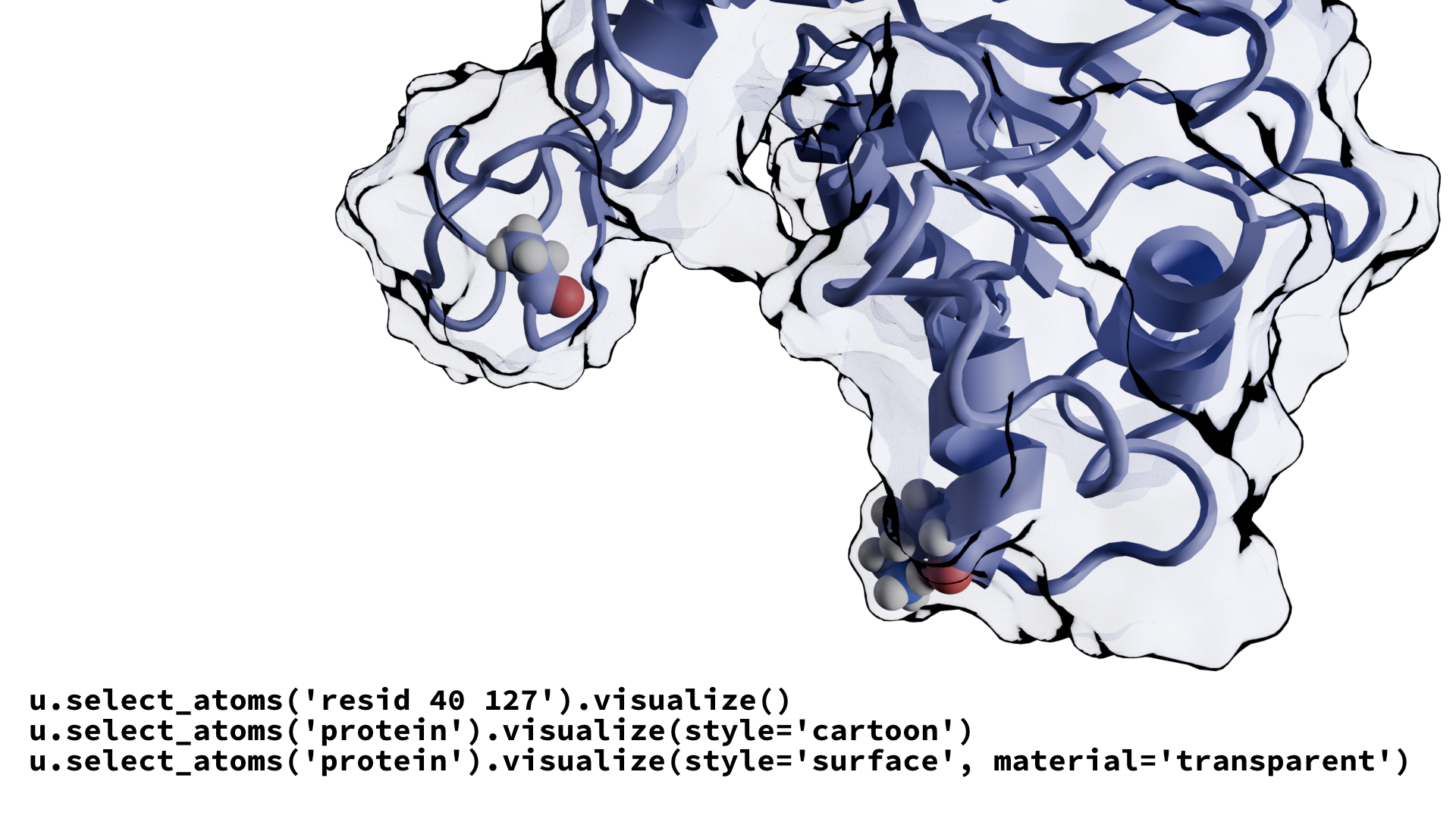
Project Overview
The development will proceed in three key stages:
-
API Development: We will create a stable API for Molecular Nodes, empowering users to automate molecular rendering with minimal effort.
-
Interactive Jupyter Integration: A Jupyter widget will be built to integrate with MDAnalysis, providing an interactive environment for controlling and rendering molecular objects directly within notebooks via Blender.
-
Advanced Visualization Tools: We will develop tools for visualizing basic geometric features and even complex analysis results from MDAnalysis.
Be Part of the Process!
We invite you to join our Discord channel to share your ideas and feedback as we build these tools. If you’d like to be a beta user, let us know—-your input will help shape the future of Molecular Nodes! Stay tuned for updates and sneak peeks of our progress.
Thank you to NumFocus for supporting this exciting project!


